MATLAB 使用共空间模式 CSP 和支持向量机 SVM 做脑电分类
在网络上找到的代码,自己做了简单的修改,跑通之后分享出来做学习交流,找不到原作者的链接了,在这里只能表示一下感谢。
共空间模式 CSP
公共空间模式算法的基本原理是利用矩阵的对角化,找到一组最优空间滤波器进行投影,使得两类信号的方差值差异最大化,从而得到具有较高区分度的特征向量。
支持向量机 SVM
支持向量机模型将样本表示为在空间中的映射的点,这样具有单一类别的样本能尽可能明显的间隔分开出来。所有这样新的样本映射到同一空间,就可以基于它们落在间隔的哪一侧来预测属于哪一类别。
函数文件目录:

先特征提取,然后分类
代码:
分类
%% 主函数
clc;
clear all;
run_FeatureExtract
disp('####### Training The SVM Classsifier ##########')
load dataCSP.mat
load labels_data_set_iii.mat
%% X为训练集, T测试集, Y训练标签
%% SVM
model = fitcsvm(X,Y);
[fpredict1,fpredict2] = predict(model,T);
fpredict=[fpredict1,fpredict2];
acc=size(find((fpredict1-y_test)==0),1)/size(y_test,1);
fprintf(strcat('acc=',num2str(acc*100),'%%'));
结果:

特征提取函数:
run_FeatureExtract:
% Extract Common Spatial Pattern (CSP) Feature
close all; clear; clc;
load dataset_BCIcomp1.mat
EEGSignals.x=x_train;
EEGSignals.y=y_train;
Y=y_train;
classLabels = unique(EEGSignals.y);
CSPMatrix = learnCSP(EEGSignals,classLabels);
nbFilterPairs = 1;
X = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs);
EEGSignals.x=x_test;
T = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs);
% save dataCSP.mat X Y T
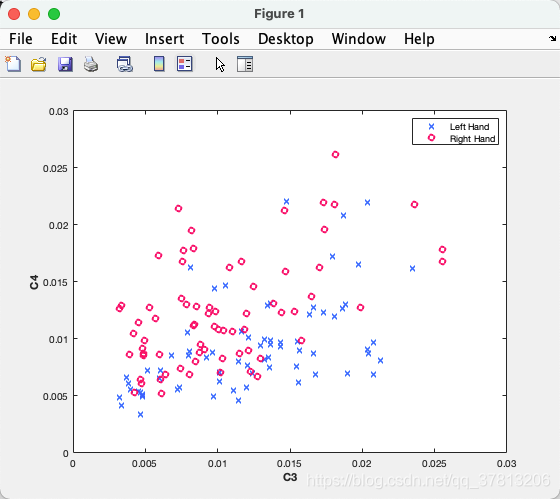
color_L = [0 102 255] ./ 255;
color_R = [255, 0, 102] ./ 255;
pos = find(Y==1);
plot(X(pos,1),X(pos,2),'x','Color',color_L,'LineWidth',2);
hold on
pos = find(Y==2);
plot(X(pos,1),X(pos,2),'o','Color',color_R,'LineWidth',2);
legend('Left Hand','Right Hand')
xlabel('C3','fontweight','bold')
ylabel('C4','fontweight','bold')
输出结果:
其中用到的learnCSP 函数和 extractCSP 函数:
learnCSP:
function CSPMatrix = learnCSP(EEGSignals,classLabels)
%
%Input:
%EEGSignals: the training EEG signals, composed of 2 classes. These signals
%are a structure such that:
% EEGSignals.x: the EEG signals as a [Ns * Nc * Nt] Matrix where
% Ns: number of EEG samples per trial
% Nc: number of channels (EEG electrodes)
% nT: number of trials
% EEGSignals.y: a [1 * Nt] vector containing the class labels for each trial
% EEGSignals.s: the sampling frequency (in Hz)
%
%Output:
%CSPMatrix: the learnt CSP filters (a [Nc*Nc] matrix with the filters as rows)
%
%See also: extractCSPFeatures
%check and initializations
nbChannels = size(EEGSignals.x,2); % 通道
nbTrials = size(EEGSignals.x,3); % 实验次数
nbClasses = length(classLabels); % 类别
if nbClasses ~= 2
disp('ERROR! CSP can only be used for two classes');
return;
end
covMatrices = cell(nbClasses,1); %the covariance matrices for each class
%% Computing the normalized covariance matrices for each trial
%% 为每个试验计算标准化的协方差矩阵。
trialCov = zeros(nbChannels,nbChannels,nbTrials);
for t=1:nbTrials
E = EEGSignals.x(:,:,t)'; %note the transpose
EE = E * E';
trialCov(:,:,t) = EE ./ trace(EE);
end
clear E;
clear EE;
%computing the covariance matrix for each class
for c=1:nbClasses
%EEGSignals.y==classLabels(c) returns the indeces corresponding to the class labels
covMatrices{c} = mean(trialCov(:,:,EEGSignals.y == classLabels(c)),3);
end
%the total covariance matrix
covTotal = covMatrices{1} + covMatrices{2};
%whitening transform of total covariance matrix
%caution: the eigenvalues are initially in increasing order注意:特征值最初是递增的
[Ut Dt] = eig(covTotal);
eigenvalues = diag(Dt);
[eigenvalues egIndex] = sort(eigenvalues, 'descend');
Ut = Ut(:,egIndex);
P = diag(sqrt(1./eigenvalues)) * Ut';
%transforming covariance matrix of first class using P
%用P变换第一类协方差矩阵
transformedCov1 = P * covMatrices{1} * P';
%EVD of the transformed covariance matrix 变换协方差矩阵的EVD
[U1 D1] = eig(transformedCov1);
eigenvalues = diag(D1);
[eigenvalues egIndex] = sort(eigenvalues, 'descend');
U1 = U1(:, egIndex);
CSPMatrix = U1' * P;
extractCSP:
function features = extractCSP(EEGSignals, CSPMatrix, nbFilterPairs)
%Input:
%EEGSignals: the EEGSignals from which extracting the CSP features. These signals
%are a structure such that:
% EEGSignals.x: the EEG signals as a [Ns * Nc * Nt] Matrix where
% Ns: number of EEG samples per trial
% Nc: number of channels (EEG electrodes)
% nT: number of trials
% EEGSignals.y: a [1 * Nt] vector containing the class labels for each trial
% EEGSignals.s: the sampling frequency (in Hz)
%CSPMatrix: the CSP projection matrix, learnt previously (see function learnCSP)
%nbFilterPairs: number of pairs of CSP filters to be used. The number of
% features extracted will be twice the value of this parameter. The
% filters selected are the one corresponding to the lowest and highest
% eigenvalues
%
%Output:
%features: the features extracted from this EEG data set
% as a [Nt * (nbFilterPairs*2 + 1)] matrix, with the class labels as the
% last column
%
%initializations
nbTrials = size(EEGSignals.x,3);
features = zeros(nbTrials, 2*nbFilterPairs+1);
Filter = CSPMatrix([1:nbFilterPairs (end-nbFilterPairs+1):end],:);
%extracting the CSP features from each trial
for t=1:nbTrials
%projecting the data onto the CSP filters
projectedTrial = Filter * EEGSignals.x(:,:,t)';
%generating the features as the log variance of the projected signals
variances = var(projectedTrial,0,2);
for f=1:length(variances)
features(t,f) = log(1+variances(f));
end
end